The first evidence that certain diseases could be caused by agents smaller than bacteria was published in 1892 by Russian microbiologist Dmitri Ivanowski, who showed that an agent capable of passing through a filter for bacteria could cause tobacco mosaic disease in plants. A few years later, Dutch microbiologist Martinus Beijerinck (1851–1931) confirmed that this agent had certain properties of living things and called it a ‘filterable virus’.
Subsequently many other diseases of plants and animals were shown to be caused by these agents, which came to be called simply viruses. The term bacteriophage (often shortened to phage) for bacterial viruses was coined in 1917 by French scientist F. d’Herelle. Today thousands of different viruses have been discovered, and a wealth of knowledge exists about their structure and mode of life. Virus-infected cells have proved extremely useful experimental systems for studying aspects of cell metabolism, including DNA replication and protein synthesis, and viruses are commonly used as vectors for cloning DNA.
Structure
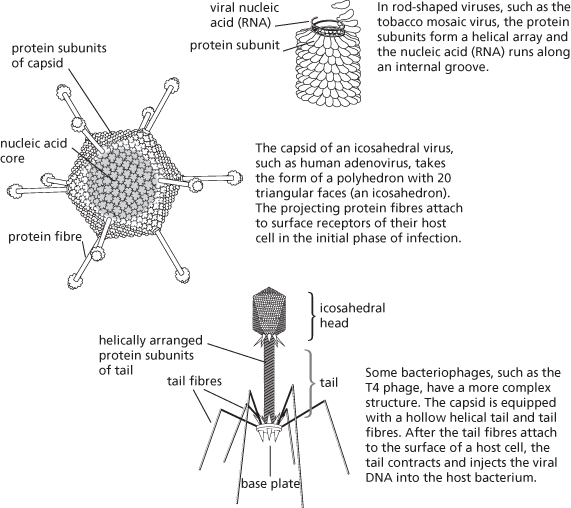
Virus particles (virions) come in various shapes and sizes, ranging from about 20 nm (e.g. parvoviruses) to 1500 nm for Pithovirus, a nucleocytoplasmic large DNA virus. Essentially they fall into two structural classes—helical and icosahedral—depending on how the viral nucleic acid and protein subunits of the capsid are arranged.
Baltimore classification
A common method of classifying viruses is based on the nature of their genetic material (DNA or RNA) and how they convert that genetic information into viral messenger RNA (mRNA). It was devised by US biologist David Baltimore (1938– ). The viral mRNA strand that is translated into viral proteins by the host cell is always denoted the positive (+) sense strand, and the template strand from which it is copied is denoted as the negative (−) sense strand.
Class
Genetic material
Genome
Replication method
Examples
I
DNA
one double-stranded DNA (dsDNA) molecule
host cell enzymes transcribe viral DNA into viral mRNA
adenoviruses, herpesviruses, poxviruses
II
DNA
one positive- or negative-sense single-stranded DNA (ssDNA) molecule
viral ssDNA is copied in host cell into dsDNA, which is copied into viral mRNA
parvoviruses
III
RNA
10–12 dsRNA molecules
negative strands of genomic RNAs copied into positive mRNAs
reoviruses (e.g. rotavirus)
IV
RNA
single positive-sense RNA strand
genomic positive strand can act as mRNA; further copies are made via synthesis of a negative RNA strand
polioviruses, togaviruses, astroviruses, caliciviruses
V
RNA
one or several positive-sense RNA strands
genomic RNA is template for viral mRNAs
orthomyxoviruses (e.g. influenza virus), paramyxoviruses, rhabdoviruses (e.g. rabies virus)
VI
RNA
two identical positive-sense RNA strands
the enzyme reverse transcriptase copies a genomic RNA strand into a single negative DNA strand, and then into dsDNA, which becomes integrated into the host genome. The host transcribes this into positive viral RNA
retroviruses (e.g. HIV)
VII
DNA
circular, part dsDNA, part
host transcribes viral DNA into mRNA, some of which ssDNA encodes viral proteins; some becomes ’pregenomic RNA’, which is copied by reverse transcriptase inside the virion into viral DNA
hepadnaviruses (e.g. hepatitis B virus)
- mechanism
- mechanism design
- mechanoreceptor
- mechanotransduction
- mechatronics
- Mecke’s test
- MECO
- meconium
- Medawar, Sir Peter Brian
- Mede
- Media Access Control address
- media convergence
- media geographies
- medial moraine
- median
- median absolute deviation
- median eye
- median filter
- median(in probability and statistics)
- median lethal dose
- median-median line
- median network
- median(of a triangle)
- median polish
- median suture