The ability of double-stranded RNA to interfere with, or suppress, the expression of a gene with a corresponding base sequence. The phenomenon occurs in many types of organisms, including plants, fungi, and animals. The RNA transcript of a noncoding RNA gene folds back on itself to form double-stranded RNA, which is normally a rarity in cells; this is cut into fragments by a ribonuclease enzyme called Dicer. There are essentially two pathways by which gene silencing is accomplished: one involves short, single-stranded microRNAs (miRNAs), encoded by the genome, which function in controlling gene expression by binding to complementary base sequences on messenger RNAs (mRNAs); the other uses short interfering RNAs (siRNAs), which target potentially ‘rogue’ mRNAs (e.g. derived from infecting viruses, or transposons) for destruction. In either case, processing by Dicer results in a short (typically 22-nucleotide) single strand of RNA being incorporated into an assemblage of proteins called the RNA-induced silencing complex (RISC). The RISC binds to the target RNA and either blocks translation (in the case of a RISC-miRNA combination) or triggers cleavage and degradation of the mRNA (with a RISC-siRNA combination). RNA interference is now used as a powerful and versatile experimental tool to suppress particular genes, as a form of knockout. See also piwi-interacting RNA.
The 1990s were remarkable for a series of advances in genetics and molecular biology, but perhaps the most amazing was the discovery of an unexpected genetic control mechanism, the phenomenon of RNA interference (RNAi).
The work of Fire and Mello
By the mid-1990s it was known that RNA is capable of inhibiting the activity of genes, particularly when the RNA base sequence matches that of the gene. But the work of US molecular geneticists Andrew Fire (1959– ) and Craig Mello (1960– ) proved to be crucial in unravelling the precise mechanism. The focus of their experiments was the nematode Caenorhabditis elegans, specifically a gene that affected muscle action. They injected worms with RNA whose base sequence matched that of the target gene. When ‘silencing’ of the target gene occurred, it caused a muscle defect in the worm and consequent twitching movements.
Key findings
• Effective silencing required double-stranded RNA molecules (dsRNAs).
• One strand of the dsRNA (the antisense strand) had to possess a base sequence complementary to that of the target gene’s mRNA and be capable of binding to it.
• Silencing was specific, affecting only the target gene mRNA.
• Silencing involved ‘mature’ mRNAs, and hence probably took place in the cytoplasm.
• The dsRNA seemed to act as some form of catalyst.
• The target mRNA disappeared from the cytoplasm, so was probably degraded.
For their discoveries concerning RNAi, Fire and Mello were jointly awarded the 2006 Nobel Prize for physiology or medicine.
Dicer and RISC
It is now established that dsRNA precursors formed in the nucleus are cut into shorter fragments (typically 21 or 22 nucleotides) in the cytoplasm by a protein called Dicer.
One strand associates with an assembly of proteins called the RNA-induced silencing complex (RISC). This binds to a complementary base sequence on its target mRNA and causes silencing.
MicroRNA (miRNA) and short interfering RNA (siRNA)
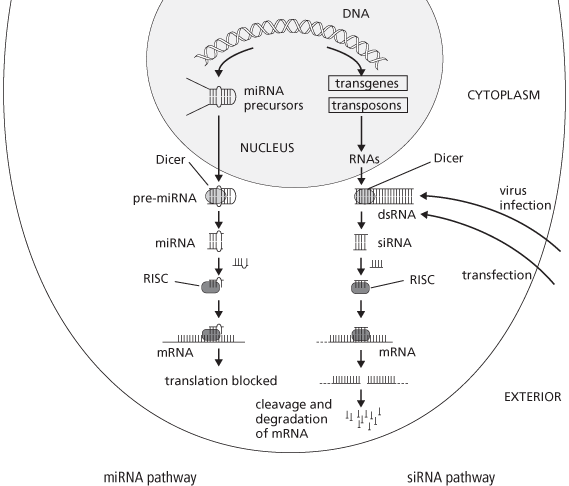
There are two basic pathways leading to RNA silencing: the miRNA pathway and the siRNA pathway, as shown in the diagram.
miRNA pathway
• Uses small RNA molecules encoded by the cell’s DNA as means of regulating gene expression.
• A double-stranded hairpin precursor (pre-miRNA) is trimmed by Dicer to form miRNA.
• The miRNA is incorporated into a RISC.
• miRNA binds imperfectly to target mRNA causing suppression of translation into protein, but not degradation of the mRNA.
siRNA pathway
• dsRNA precursors originate from various sources, including virus infection, introduced transgenes, and transposons.
• Dicer trims precursors to form siRNAs, which are incorporated in RISCs.
• siRNA–RISC binds to its target mRNA sequence completely, triggering cleavage and degradation of the mRNA.
Functions of RNAi
• Regulatory mechanism for an estimated 50% of all protein-coding genes (in mammals).
• Helps protect cells against certain viruses by targeting viral RNA for destruction.
• Helps to silence potentially disruptive transposons in the genome by destroying RNA copies arising from transposon replication.
Applications
RNAi is a precise and efficient tool for knockout of specific genes when studying gene function in experimental organisms (although, unlike genome editing, its effects are temporary). It also has potential for new forms of targeted but reversible gene therapy.
- command and control
- command and service module
- command control language
- command control program
- command economy
- commander
- command file
- command language
- command-line interface
- command-loss timer
- command module
- command sequence
- command system
- commensalism
- commensurability
- commensurable
- commensurate lattice
- comment
- commerce bot
- commerce server
- commerce service provider
- commercial agriculture
- commercial bank
- commercial bill
- commercial off-the-shelf