A living cell makes and uses thousands of different RNAs and proteins to perform all its vital functions. Real insight into cellular metabolism requires knowledge of which components are being expressed by different cell types, for example during development, under different physiological conditions, or in disease states, and how their concentrations change over time.
Microarrays were developed in the 1990s for the automated analysis of thousands of different biomolecules simultaneously. They have enabled researchers to study the function of genes and their products (RNAs and proteins) on a genome-wide basis—the field of functional genomics. These techniques generate massive amounts of data, and computerized handling, analysis, and presentation of the data are crucial for valid interpretation.
Examples of microarrays
DNA microarrays (‘gene chips’ or ‘DNA chips’)
Contain short oligonucleotides or larger DNA fragments that hybridize with complementary DNA or RNA sequences. Uses include:
• following expression patterns of whole genomes over time;
• identifying chemical changes throughout a cell’s chromosomal DNA;
• identifying and measuring minute deletions and duplications of DNA, e.g.in cancer cells.
Protein microarrays
Contain microsamples of purified proteins.
Used to determine protein-protein interactions at a cellular scale.
Tissue microarrays
Contain small thin sections of tissue taken from different tissue locations or pathology specimens.
Used to identify distribution of specific proteins throughout a tissue sample.
Chemical microarrays
Contain chemical compounds.
Used, for example, to select potential drugs with particular chemical binding properties.
Preparation of DNA microarrays
There are two basic types of DNA microarrays.
In situ oligonucleotide arrays (DNA chips)
DNA oligonucleotides (20–25 nucleotides) are assembled in situ on a quartz wafer or glass slide, using a photolithography process similar to that used to make microprocessor chips.
A single chip may contain up to a million specified oligonucleotides at regularly spaced sites called addresses, or ‘features’.
Poor or incorrect hybridization leads to signal ‘noise’.
Spotted microarrays
Based on the principle of the dot-blot technique with complementary DNA obtained from clones of the genes of interest.
Assembled by spotting minute amounts of denatured DNA onto a coated glass microscope slide, so that the DNA dries and becomes attached to the slide.
Contain large DNA fragments (hundreds to thousands of base pairs long).
Up to 20 000 features per slide.
Cheaper and easier to make—can be made ‘in house’ by researchers.
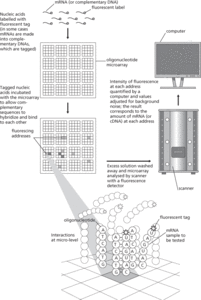
Measuring mRNAs using an oligonucleotide microarray
- molar
- molar conductivity
- molar density
- molar flow rate
- molar flux
- molar heat capacity
- molarity
- molar latent heat
- molar volume
- molasse
- Molasse Basin
- moldavite
- moldic porosity
- Moldova
- mole
- mole balance
- molecular beam
- molecular-beam epitaxy
- molecular biology
- molecular chaperone
- molecular clock
- molecular cloud
- molecular crystal
- molecular diameter
- molecular diffusion